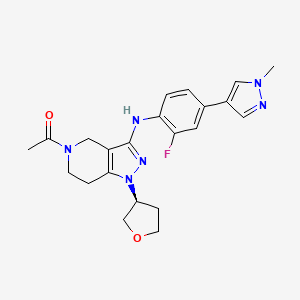
GNE-272
(S)-1-(3-((2-fluoro-4-(1-methyl-1H-pyrazol-4- yl)phenyl)amino)-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin- 5(4H)-yl)ethanone
1-[3-[2-fluoro-4-(1-methylpyrazol-4-yl)anilino]-1-[(3S)-oxolan-3-yl]-6,7-dihydro-4H-pyrazolo[4,3-c]pyridin-5-yl]ethanone
CAS 1936428-93-1
| Molecular Formula: | C22H25FN6O2 |
|---|---|
| Molecular Weight: | 424.471303 g/mol |
GENENTECH, INC. [US/US]; 1 DNA Way South San Francisco, California 94080-4990 (US).
CONSTELLATION PHARMACEUTICALS, INC. [US/US]; 215 First Street Suite 200 Cambridge, Massachusetts 02142 (US)
ROMERO, F. Anthony; (US).
MAGNUSON, Steven; (US).
PASTOR, Richard; (US).
TSUI, Vickie Hsiao-Wei; (US).
MURRAY, Jeremy; (US).
CRAWFORD, Terry; (US).
ALBRECHT, Brian, K.; (US).
COTE, Alexandre; (US).
TAYLOR, Alexander, M.; (US).
LAI, Kwong Wah; (CN).
CHEN, Kevin, X.; (CN).
BRONNER, Sarah; (US).
ADLER, Marc; (US).
EGEN, Jackson; (US).
LIAO, Jiangpeng; (CN).
WANG, Fei; (CN).
CYR, Patrick; (US).
ZHU, Bing-Yan; (US).
KAUDER, Steven; (US)
Chromatin is a complex combination of DNA and protein that makes up chromosomes. It is found inside the nuclei of eukaryotic cells and is divided between heterochromatin (condensed) and euchromatin (extended) forms. The major components of chromatin are DNA and proteins. Histones are the chief protein components of chromatin, acting as spools around which DNA winds. The functions of chromatin are to package DNA into a smaller volume to fit in the cell, to strengthen the DNA to allow mitosis and meiosis, and to serve as a mechanism to control expression and DNA replication. The chromatin structure is controlled by a series of post-translational modifications to histone proteins, notably histones H3 and H4, and most commonly within the “histone tails” which extend beyond the core nucleosome structure. Histone tails tend to be free for protein-protein interaction and are also the portion of the histone most prone to post-translational modification. These modifications include acetylation, methylation, phosphorylation, ubiquitinylation, and SUMOylation. These epigenetic marks are written and erased by specific enzymes that place the tags on specific residues within the histone tail, thereby forming an epigenetic code, which is then interpreted by the cell to allow gene specific regulation of chromatin structure and thereby transcription.
Of all classes of proteins, histones are amongst the most susceptible to post-translational modification. Histone modifications are dynamic, as they can be added or removed in response to specific stimuli, and these modifications direct both structural changes to chromatin and alterations in gene transcription. Distinct classes of enzymes, namely histone acetyltransferases (HATs) and histone deacetylases (HDACs), acetylate or de-acetylate specific histone lysine residues (Struhl K., Genes Dev., 1989, 12, 5, 599-606).
Bromodomains, which are approximately 1 10 amino acids long, are found in a large number of chromatin-associated proteins and have been identified in approximately 70 human proteins, often adjacent to other protein motifs (Jeanmougin F., et al., Trends Biochem. Sc , 1997, 22, 5, 151-153; and Tamkun J.W., et al., Cell, 1992, 7, 3, 561-572).
Interactions between bromodomains and modified histones may be an important mechanism underlying chromatin structural changes and gene regulation. Bromodomain-containing proteins have been implicated in disease processes including cancer, inflammation and viral replication. See, e.g., Prinjha et al,, Trends Pharm. Sci., 33(3):146-153 (2012) and Muller et al , Expert Rev. , 13 (29): 1 -20 (September 201 1 ).
Cell-type specificity and proper tissue functionality requires the tight control of distinct transcriptional programs that are intimately influenced by their environment.
Alterations to this transcriptional homeostasis are directly associated with numerous disease states, most notably cancer, immuno-inflammation, neurological disorders, and metabolic diseases. Bromodomains reside within key chromatin modifying complexes that serve to control distinctive disease-associated transcriptional pathways. This is highlighted by the observation that mutations in bromodomain-containing proteins are linked to cancer, as well as immune and neurologic dysfunction. Hence, the selective inhibition of bromodomains across a specific family, such as the selective inhibition of a bromodomain of CBP/EP300, creates varied opportunities as novel therapeutic agents in human dysfunction.
There is a need for treatments for cancer, immunological disorders, and other
CBP/EP300 bromodomain related diseases.
PATENT
Scheme 1


Scheme 2

Scheme 3

Scheme 4

General procedure for Intermediates A & B

Intermediate A

Intermediate
General procedure for Intermediates F & G

Intermediate F

Intermediate G
Step 1:
(R)-tetrahydrofuran-3-yI methanesulfonate

To a solution of (^)-tetrahydrofuran-3-ol (25 g, 253.7 mmol) in DCM (250 mL) at 0 °C was added triethylamine (86 g, 851.2 mmol) and mesyl chloride (39 g, 340.48 mmol) dropwise. The mixture was stirred at room temperature for 12 h. The reaction was quenched with water (100 mL) and extracted with DCM (100 mL x 2). The combined organic layers were dried over anhydrous Na2S04, filtered and concentrated in vacuo to give the title compound (47 g, 99%) as a brown oil. Ή NMR (400 MHz, CDC13) δ 5.35 – 5.27 (m, 1H), 4.05 – 3.83 (m, 4H), 3.04 (s, 3 H), 2.28 – 2.20 (m, 2 H).
Step 2:
(S)-tert-butyl 3-bromo-l-(tetrahydrofuran-3-yI)-6,7-dihydro-li/-pyrazolo[43- c] pyridine-5(4H)-carboxylate

To a solution of tert-butyl 3-bromo-6,7-dihydro-lH-pyrazolo[4,3-c]pyridine-5(4H)-carboxylate (Intermediate A, 24.8 g, 82 mmol) in DMF (200 mL) was added Cs2C03 (79 g, 246 mmol) and (/?)-tetrahydrofuran-3-yl methanesulfonate (17.4 g, 98 mmol). The mixture was heated to 80 °C for 12 h. After cooling the reaction to room temperature, the mixture was concentrated in vacuo. The crude residue was purified by silica gel chromatography
(petroleum ether / EtOAc = from 10 : 1 to 3 : 1) to give the title compound (Intermediate F, 50 g, 71 %) as a yellow oil. Ή NMR (400 MHz, DMSO-i ) δ 4.97 – 4.78 (m, 1H), 4.13 (s, 2H), 3.98 – 3.86 (m, 2H), 3.81 – 3.67 (m, 2H), 3.56 (t, J= 5.6 Hz, 2H), 2.68 (t, J= 5.6 Hz, 2H), 2.33 – 2.08 (m, 2H), 1.38 (s, 9H).
Step 3:
(5)-l-(3-bromo-l-(tetrahydrofuran-3-yl)-6,7-dihydro-lH-pyrazoIo[4,3-c]pyridin-5(4//)- yl)ethanone

To a solution of (S)-tert-buty\ 3-bromo- 1 -(tetrahydrofuran-3-yl)-6,7-dihydro-lH-pyrazolo [4,3 -c]pyridine-5(4H)-carboxy late (29 g, 78 mmol) in DCM (300 mL) was added trifluroacetic acid (70 mL) dropwise. The mixture was stirred at room temperature for 2 h. The solvent was concentrated in vacuo and the crude residue was re -dissolved in DMF (100 mL). The mixture was cooled to 0 °C before triethylamine (30 g, 156 mmol) and acetic anhydride (8.7 g, 86 mmol) were added dropwise. The mixture was stirred at room temperature for an additional 2 h. The reaction was quenched with water (200 mL) at 0 °C and extracted with EtOAc (150 mL x 3). The combined organic layers were dried over anhydrous Na2S0 , filtered and concentrated in vacuo. The crude residue was purified by silica gel chromatography (DCM / MeOH = 30 : 1) to give the title compound (Intermediate G, 21.3 g, 87%) as a white solid. lH NMR (400 MHz, CDC13) δ 4.78 – 4.67 (m, 1H), 4.45 -4.29 (m, 2H), 4.15 – 4.06 (m, 2H), 3.96 – 3.92 (m, 2H), 3.88 – 3.70 (m, 2H), 2.71 – 2.67 (m, 2H), 2.38 – 2.34 (m, 2H), 2.16 (s, 3H).
PATENT

| Example 300 | 1-[3-[2-fluoro-4-(1-methylpyrazol-4- yl)anilino]-1-[(3S)-tetrahydrofuran-3- yl]-6,7-dihydro-4H-pyrazolo[4,3- c]pyridin-5-yl]ethanone |
1H NMR (400 MHz, DMSO- d6) δ 8.03 (s, 1H), 7.83-7.68 (m, 3H), 7.36-7.33 (m, 1H), 7.32-7.21 (m, 1H), 4.88- 4.84 (m, 1H), 4.40-4.33 (m, 2H), 4.03-3.99 (m, 2H), 3.84- 3.67 (m, 7H), 2.79-2.64 (m, 2H), 2.26-2.21 (m, 2H), 2.08-2.05 (m, 3H) | 425 |
General Procedure for Intermediates F & G
Step 1
(R)-tetrahydrofuran-3-yl methanesulfonate
Step 2
(S)-tert-butyl 3-bromo-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridine-5(4H)-carboxylate
Step 3
(S)-1-(3-bromo-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone
OTHER ISOMER

| Example 299 | 1-[3-[2-fluoro-4-(1-methylpyrazol-4- yl)anilino]-1-[(3R)-tetrahydrofuran-3- yl]-6,7-dihydro-4H-pyrazolo[4,3- c]pyridin-5-yl]ethanone |
1H NMR (400 MHz, DMSO- d6) δ 8.03 (s, 1H), 7.83-7.67 (m, 3H), 7.39-7.34 (m, 1H), 7.26-7.21 (m, 1H), 4.87- 4.77 (m, 1H), 4.41-4.34 (m, 2H), 4.02-3.97 (m, 2H), 3.83 (s, 3H), 3.81-3.67 (m, 4H), 2.77-2.66 (m, 2H), 2.26- 2.22 (m, 2H), 2.08-2.05 (m, 3H) | 425 |
PAPER

The single bromodomain of the closely related transcriptional regulators CBP/EP300 is a target of much recent interest in cancer and immune system regulation. A co-crystal structure of a ligand-efficient screening hit and the CBP bromodomain guided initial design targeting the LPF shelf, ZA loop, and acetylated lysine binding regions. Structure–activity relationship studies allowed us to identify a more potent analogue. Optimization of permeability and microsomal stability and subsequent improvement of mouse hepatocyte stability afforded 59 (GNE-272, TR-FRET IC50 = 0.02 μM, BRET IC50 = 0.41 μM, BRD4(1) IC50 = 13 μM) that retained the best balance of cell potency, selectivity, and in vivo PK. Compound 59 showed a marked antiproliferative effect in hematologic cancer cell lines and modulates MYC expression in vivo that corresponds with antitumor activity in an AML tumor model.
Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300
UNDESIRED R ISOMER
In a similar procedure to59, the title compound was prepared from (S)-tetrahydrofuran-3-yl
methanesulfonate and purified by Prep-TLC (DCM / MeOH = 15 : 1) to give the title
compound as a light yellow solid.
1H NMR (400 MHz, CDCl3) δ 7.76–7. 42 (m,1H), 7.68 (s, 1H), 7.53 (s, 1H), 7.20–7.12 (m, 2H), 5.86–5.77 (m, 1H), 4.79–4.69 (m, 1H),4.47–4.29 (m, 2H), 4.25–4.08 (m, 2H), 4.06–3.72 (m, 4H), 3.99 (s, 3H), 2.76–2.65 (m, 2H),
2.49–2.28 (m, 2H), 2.25–2.12 (m, 3H).
13C NMR (100 MHz, CDCl3) δ 169.81, 169.36,151.71 (d, J = 238.9 Hz), 145.51, 144.64, 137.83, 136.32, 135.89, 126.35, 121.41, 116.44 (d,J = 26.0 Hz), 111.88, 103.09 (d, J = 24.0 Hz), 71.94, 68.10, 57.65, 43.24, 42.24, 39.02, 37.83,32.49, 22.01.
LCMS M/Z (M+H) 425.
[α]27D +8.8 (c 0.78, CHCl3, 99% ee).
DESIRED S ISOMER
(S)-1-(3-((2-fluoro-4-(1-methyl-1H-pyrazol-4- yl)phenyl)amino)-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin- 5(4H)-yl)ethanone
aReagents and conditions: (a) 4-bromo-2-fluoro-1-isothiocyanato-benzene, KOtBu, THF, rt (b) CH3I, 40 °C, 51%; (c) hydrazine monohydrate, EtOH, 85 °C; 96%; (d) 1-methyl-4-(4,4,5,5- tetramethyl-1,3,2-dioxaborolan-2-yl)pyrazole, dioxane / water, Na2CO3, Pd(dppf)Cl2, 100 °C, 63%; (e) (R)-tetrahydrofuran-3-yl methanesulfonate, Cs2CO3, DMF, 90 oC, 42%.
The crude residue was purified by silica gel chromatography (DCM / MeOH = 100:1) to give (S)-1-(3-((2-fluoro-4-(1- methyl-1H-pyrazol-4-yl)phenyl)amino)-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1Hpyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone as a light yellow solid.
1H NMR (400 MHz, CDCl3) δ 7.76–7.72 (m, 1H), 7.68 (s, 1H), 7.53 (s, 1H), 7.20–7.12 (m, 2H), 5.86–5.77 (m, 1H), 4.79–4.69 (m, 1H), 4.47–4.29 (m, 2H), 4.25–4.08 (m, 2H), 4.06– 3.72 (m, 4H), 3.99 (s, 3H), 2.76–2.65 (m, 2H), 2.49–2.28 (m, 2H), 2.25–2.12 (m, 3H).
13C NMR (100 MHz, CDCl3) δ 169.8, 169.4, 151.7 (d, J = 238.9 Hz), 145.5, 144.64, 137.83, 136.3, 135.9, 126.4, 121.4, 116.4 (d, J = 26.0 Hz), 111.9, 103.1 (d, J = 24.0 Hz), 71.9, 68.1, 57.7, 43.2, 42.2, 39.0, 37.8, 32.5, 22.0.
LCMS m/z (M+H) 425.
[α]27 D -11.0 (c 1.0, CHCl3, 99% ee).
HRMS m/z 425.2093 (M + H+ , C22H25FN6O2, requires 425.2057).
//////////GNE-272, Genentech, CBP, EP300, cancer, immune system regulation, 1936428-93-1, WO 2016086200
[H][C@@]1(CCOC1)N1N=C(NC2=C(F)C=C(C=C2)C2=CN(C)N=C2)C2=C1CCN(C2)C(C)=O